Genome editing or gene editing tools bring along with them a promise for a better future as they give scientists the ability to alter DNA. Further, due to the growing use of technologies for gene therapy, the global market of Genetic Engineering is presumed to expand at an impressive 14.5% CAGR from 2018 through 2027.
Various tools are available in cell engineering, like CRISPR, ZNF, TALEN, and Meganucleases for gene editing. However, out of all these, CRISPR has proved to be the most popular. It has created a lot of buzz in the scientific community because it is faster and cheaper, and its results are more accurate than other existing genome editing methods. CRISPR promises many potential applications, including correcting genetic defects, treating and preventing the spread of diseases, and improving crops.
What is CRISPR?
CRISPR/Cas9 is a versatile genome editing technology that is widely used for creating genetically modified organisms as well as in preclinical research of genetic disorders. However, it’s not a perfect system. Occasionally cutting at the wrong place and not working as intended leaves scientists scratching their heads.
Well, now there are a few solutions investigated by researchers that can assist CRISPR in snipping out more than 90 percent of all genetic diseases.
How does CRISPR Cas9 work?
CRISPR utilizes an enzyme called Cas9 that uses an RNA molecule to guide navigating toward its targeted DNA. It then edits or modifies the DNA, which can deactivate genes or insert the desired sequence to achieve a behavior. Its most promising application is genetically modifying cells to overcome genetic defects and their potential to cure diseases like cancer.
Problems with CRISPR
There are certain problems with CRISPR, such as unwanted off-target mutations. It works by cutting the double-stranded DNA at precise locations in the genome. When the cell’s natural repair process takes over, it can cause damage. Further, it could create unwanted off-target mutations where the modified DNA is inserted at the cut site.
That’s a problem. But as they say, if there’s a problem, there are solutions too. One just needs to look harder.
Curious to gauge the available solutions, our research team started on the task of finding all the research done in the domain that could solve the aforementioned problem. After a bit of digging later, we found some results from patent documents that are capable of solving this problem.
 Solutions that can make CRISPR a better tool
Solutions that can make CRISPR a better tool
We analyzed research papers and patent applications to find the solutions researchers are using to make CRISPR better. Below are the top results that we found during the research.
Modification in the CRISPR enzymes [US20190032036A1]
Researchers at MIT filed a patent for a solution to modify the CRISPR enzyme to solve the off-target effect. Because DNA is negatively charged, we know that it binds itself to a groove in Cas9 protein which is positively charged. Zhang replaced some of the positively charged amino acids with neutral ones to decrease the binding of “off-target” sequences.
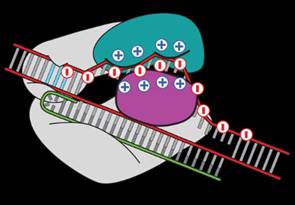 Zhang’s team found that mutations in three amino acids dramatically reduced “off-target” cuts. They have created a newly engineered enzyme – “enhanced” S. pyogenes Cas9, or eSpCas9, which will be useful for specific genome editing applications.
Zhang’s team found that mutations in three amino acids dramatically reduced “off-target” cuts. They have created a newly engineered enzyme – “enhanced” S. pyogenes Cas9, or eSpCas9, which will be useful for specific genome editing applications.
Implementation of Second guide RNA [US20170247671A1]
During a CRISPR-based edit, a strand of molecules called a guide RNA leads the DNA-slicing protein Cas9 to the section of DNA targeted for editing. Once the guide RNA binds to the DNA, Cas9 cuts so that new DNA can be inserted or deleted.
Chong Wing Yung and Andrew Kennedy of Agilent Technologies suggested that to prevent off-target binding, an additional second guide RNA can be implied that has a blocking guide sequence that is complementary to an off-target nucleic acid sequence.
In-Silico techniques [US10354746B2]
There could be quite a bit of randomness in what happens during these CRISPR edits, and that randomness can potentially create unexpected outcomes. To minimize them, a team at Georgia is working on an algorithm that takes in data and identifies cleavage locations of Cas9 nuclease and selects the nuclease having the fewest off-target cleavage locations.
Digenome sequencing method for detecting off-target effect [US20190153530A1]
To validate off-target sites, an in vitro Cas9-digested whole-genome sequencing technique, Digenome-seq, is developed by scientists at the Institute for Basic Science in Seoul, Korea. This in vitro method yields sequences that can be computationally identified to profile genome-wide Cas9 off-target effects in human cells. Digenome-seq is a robust, sensitive, unbiased, and cost-effective method for profiling genome-wide off-target effects of programmable nucleases, including Cas9.
An upgraded version of CRISPR – CRISPR Prime
Prime editing is created by scientist David Liu of the Broad Institute of MIT and Harvard and postdoctoral fellow Andrew V. Anzalone. Interestingly, the technique can potentially fix more than 90% of known genetic diseases, including Tay-Sachs disease.
This versatile and precise genome editing method works by directly writing new genetic information into a specified DNA site using a catalytically impaired Cas9 endonuclease fused to an engineered reverse transcriptase, programmed with a prime editing guide RNA (pegRNA) that both specify the target site and encodes the desired edit.
CRISPR prime also offers the versatile function of multi-letter base-editing [Can tackle four DNA letters mutation genetic disorders like Tay-Sachs] with minimized DNA damage, less off-target effect, and efficient DNA healing.
A guide RNA called pegRNA guides the Cas9 enzyme to snip out only a single strand of DNA and prevents the double-strand breaks, which can induce unintended disruptions. After that, the reverse transcriptase enzyme directly copies the edited genetic information contained in the pegRNA to the targeted genomic site. This helps generate cells, which will help patients to recover and heal, as well as help develop new vaccines against deadly diseases.
Conclusion
Researchers in R&D spend a lot of time studying research papers, but searching within papers does not always yield similar output as we get in patents. As a matter of fact, in the majority of cases, search within patents is not fully explored, which has its repercussions. To ensure a comprehensive search, it’s recommended that researchers combine journal research with an expert patent search so that multiple new solutions can be identified.
BTW, if you are researching in the genetic engineering domain and looking for solutions to problems like:
- How to improve the delivery-related problems of genetic engineering?
- How to improve the immunosuppression and immune activation problem of mammalian cell engineering?
- How to improve over activation of CAR T cells?
and others, you should get in touch. We have the solutions; all you need to do is ask.
Authored by: Anjali Gaikwad, Research Analyst, Life Science.